Table of Contents
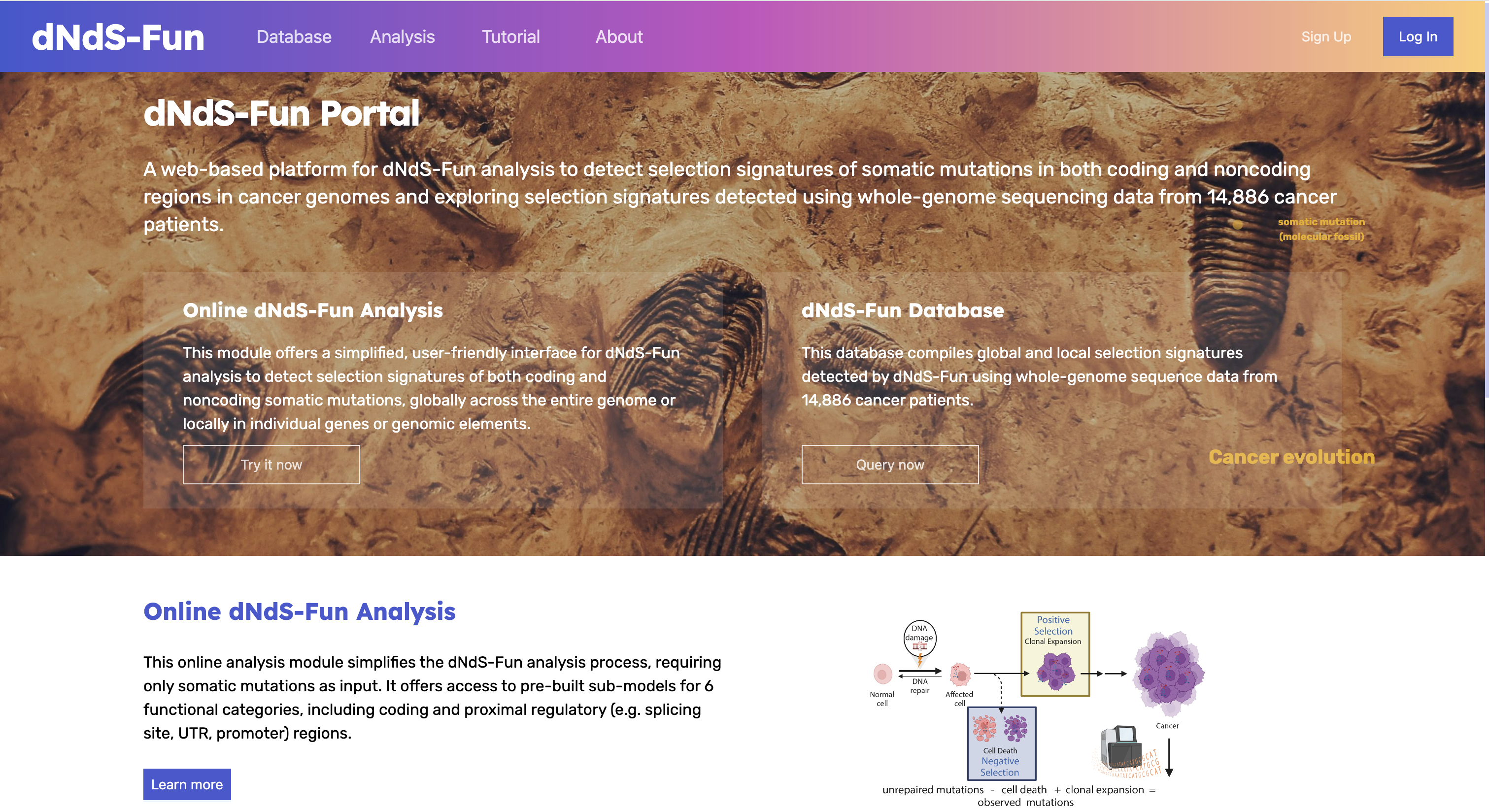
dNdS-Fun is a generalized framework that extends the classical dN/dS methodology, specifically dNdScv, to detect and quantify selection signatures on both coding and noncoding somatic mutations in cancer genomes. By integrating genome-wide functional impact scores, dNdS-Fun allows for the identification of positive and negative selection of both coding and noncoding mutations at global (genome-wide) and local (gene or element-specific) scales.
Our Portal #
Our group #
Contact #
If you have any questions about dNdS-Fun, please feel free to open an issue on the GitHub repository or contact us via email at jian.yang@westlake.edu.cn.
Citation #
Zheng M†, Sun X†, Hou J, Guo M, Liu X, Yang W, Yang J (2024) Characterizing selection signatures in coding and noncoding regions of 14,886 cancer genomes. Submitted. († Equal contribution).
Acknowledgements #
Mengyue Zheng (methods, tool development, data analysis, and documentation), Junren Hou (online tool development and maintenance), Weixi Sun (supervision, methods), and Jian Yang (supervision, methods, documentation, and maintenance). We thank the Westlake University High-Performance Computing Centre for hosting the online tool and portal.